Description of running Matlab source
code for cancer accuracy analysis using PCA and ICA
By Nasser Abbasi, June 3, 2007
Code used in conference paper
There is only one m file needed to run the PCA accuracy. Here is the Matlab source code nma_PCAaccuracy.m and here it is in HTML format html\nma_PCAaccuracy.html
The matlab file requires that you have the following 2 text files in the same folder as this file
Note: The file nma_PCAaccuracy.m makes calls to some functions in Matlab stats toolbox. These are the additional files needed:
If you do not have access to these files, I've put them in this zip file support_matlab_files.zip
Download this file and unzip its content in the same folder.
Now when you run the file nma_PCAaccuracy.m, the PCA cancer accuracy results will be printed on the console.
This Matlab file was written by Dr Lee to do more statistics. I include here as reference in case it is needed. stats.m
This Matlab file I wrote initially when I was working to use Matlab bioinformatics toolbox to download data directly from GEO databases. This file requires Matlab bioinformatics 2.5 toolbox. It will download all the files needed and used by Chen original paper and used by this project. But we did not end up using this data anyway and switched to using the original data since it was already cleaned up. May be in the future I can revisit this. nma_microarray_main.m To run this file, you need to have the following 2 text files in the same folder. It tells it which files to download from the internet. GSM_names_normal_liver.txt and GSM_names_tumor_liver.txt
Code for PCA followed by ICA
This code below was written by Michael Vodhanel as far as I know and may be by others as well.
This code is for a different project than the one I worked on above. Where PCA is used first to generate the 4 most dominant eigenmodes, followed by feeding these to ICA. This project is not fully complete, but I have fixed the PCA code in the file POD_Modes.m so that now the sign of the dominant PHI's are correct as we did with the paper version of the code above.
All of the source is in this zip file PCA_and_ICA_project_june_8_2007.zip [9 MB]
To run the code, execute this Matlab script BladderGUI.m which start a GUI interface.
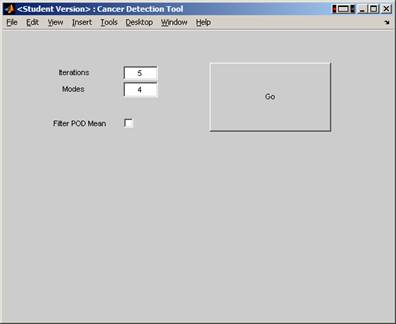
Then click on the button to run it (after checking 'Filter POD Mean' button.
The
above GUI will call the function Bladder_ICA_Modes_Itt.m
Which will call POD_Modes.m to perform PCA. Then the function ICA.m is called.